Note
Go to the end to download the full example code
XOR classification
A simple example for getting started with emlearn.
Will train a RandomForestClassifier model on a XOR dataset, generate C code for this model using emlearn Python package, load this model in C and make predictions using it.
import os.path
import emlearn
import numpy
import pandas
import seaborn
import matplotlib.pyplot as plt
try:
# When executed as regular .py script
here = os.path.dirname(__file__)
except NameError:
# When executed as Jupyter notebook / Sphinx Gallery
here = os.getcwd()
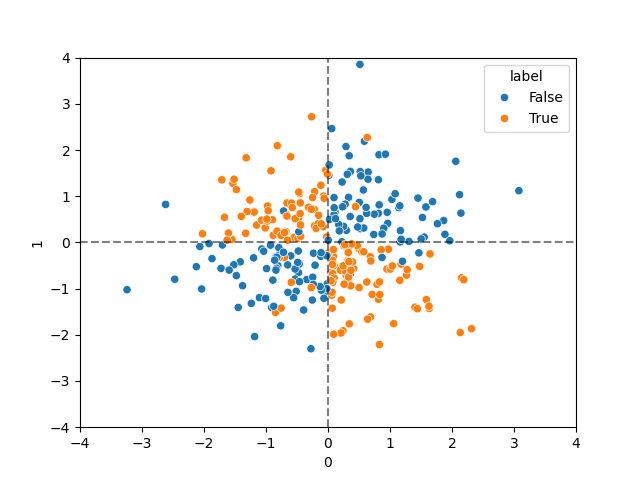
Create dataset
The XOR problem is a very simple example of a dataset which is not linearly separable.
def make_noisy_xor(seed=42):
xx, yy = numpy.meshgrid(numpy.linspace(-3, 3, 500),
numpy.linspace(-3, 3, 500))
rng = numpy.random.RandomState(seed)
X = rng.randn(300, 2)
y = numpy.logical_xor(X[:, 0] > 0, X[:, 1] > 0)
# Add some noise
flip = rng.randint(300, size=15)
y[flip] = ~y[flip]
df = pandas.DataFrame(X)
df['label'] = y
return df
def dataset_split_random(data, val_size=0.25, test_size=0.25, random_state=3, column='split'):
"""
Split DataFrame into 3 non-overlapping parts: train,val,test with specified proportions
Returns a new DataFrame with the rows marked by the assigned split in @column
"""
train_size = (1.0 - val_size - test_size)
from sklearn.model_selection import train_test_split
train_val_idx, test_idx = train_test_split(data.index, test_size=test_size, random_state=random_state)
val_ratio = (val_size / (val_size+train_size))
train_idx, val_idx = train_test_split(train_val_idx, test_size=val_ratio, random_state=random_state)
data = data.copy()
data.loc[train_idx, column] = 'train'
data.loc[val_idx, column] = 'val'
data.loc[test_idx, column] = 'test'
return data
dataset = make_noisy_xor()
dataset = dataset_split_random(dataset, test_size=0.10).set_index('split')
# Plot the dataset
ax = seaborn.scatterplot(data=dataset, x=0, y=1, hue='label')
ax.axvline(0.0, ls='--', alpha=0.5, color='black')
ax.axhline(0.0, ls='--', alpha=0.5, color='black')
ax.set_xlim(-4.0, +4.0)
ax.set_ylim(-4.0, +4.0)
# Show colums of the data
print(dataset.head(5))
0 1 label
split
val 0.496714 -0.138264 True
val 0.647689 1.523030 False
val -0.234153 -0.234137 False
train 1.579213 0.767435 False
train -0.469474 0.542560 True
Train ML model
Usin the standard process with scikit-learn
def train_model(dataset, seed=42):
from sklearn.ensemble import RandomForestClassifier
#feature_columns =
X_train = dataset.loc['train', [0, 1]]
Y_train = dataset.loc['train', 'label']
model = RandomForestClassifier(n_estimators=10, max_depth=5, random_state=seed)
model.fit(X_train, Y_train)
return model
model = train_model(dataset)
Convert model to C using emlearn
def convert_model(model):
model_filename = os.path.join(here, 'xor_model.h')
cmodel = emlearn.convert(model)
code = cmodel.save(file=model_filename, name='xor')
assert os.path.exists(model_filename)
print(f"Generated {model_filename}")
convert_model(model)
Generated /home/docs/checkouts/readthedocs.org/user_builds/emlearn/checkouts/latest/examples/xor_model.h
Use generated C model to make predictions
xor.c : Executable that takes features as commandline arguments, and prints the predicted class to stdout
#include "xor_model.h" // emlearn generated model
#include <stdio.h> // printf
#include <stdlib.h> // stdod
int
main(int argc, const char *argv[])
{
if (argc != 3) {
return -1;
}
const float a = strtod(argv[1], NULL);
const float b = strtod(argv[2], NULL);
const float features[] = { a, b };
const int out = xor_predict(features, 2);
if (out < 0) {
return out; // error
} else {
printf("%d\n", out);
}
}
# Python wrapper for the C executable
# calls the C program as a subprocess to run the model
def predict(bin_path, X, verbose=1):
import subprocess
def predict_one(x):
args = [ bin_path, str(x[0]), str(x[1]) ]
out = subprocess.check_output(args)
cls = int(out)
if verbose > 0:
print(f"run xor in1={x[0]:+.2f} in2={x[1]:+.2f} out={cls} ")
return cls
y = [ predict_one(x) for x in numpy.array(X) ]
return numpy.array(y)
def evaluate_model(dataset):
# Compile the xor.c example program
out_dir = './examples'
src_path = os.path.join(here, 'xor.c')
include_dirs = [ emlearn.includedir ]
bin_path = emlearn.common.compile_executable(src_path, out_dir, include_dirs=include_dirs)
print('Compiled C excutable', bin_path)
# Make predictions on dataset
X_test = dataset.loc['test', [0, 1]]
Y_test = dataset.loc['test', 'label']
y_pred_c = predict(bin_path, X_test)
y_pred_py = model.predict(X_test)
# Compute scores using converted C model, and original Python model
import sklearn.metrics
f1_score_c = sklearn.metrics.f1_score(Y_test, y_pred_c)
f1_score_py = sklearn.metrics.f1_score(Y_test, y_pred_py)
print(f'\nF1-score python={f1_score_py} c={f1_score_c}')
evaluate_model(dataset)
Compiled C excutable ./examples/main
run xor in1=-0.46 in2=-0.47 out=0
run xor in1=-0.01 in2=-1.06 out=0
run xor in1=+0.32 in2=-0.39 out=1
run xor in1=-1.11 in2=-1.20 out=0
run xor in1=+1.40 in2=-1.40 out=1
run xor in1=-1.55 in2=+0.07 out=1
run xor in1=-0.92 in2=+1.55 out=1
run xor in1=+1.87 in2=+0.47 out=0
run xor in1=+0.82 in2=+1.90 out=0
run xor in1=-0.25 in2=-0.75 out=0
run xor in1=+0.52 in2=+0.51 out=0
run xor in1=-0.49 in2=+0.08 out=1
run xor in1=-0.73 in2=+0.22 out=1
run xor in1=-2.03 in2=+0.19 out=1
run xor in1=-0.44 in2=+0.13 out=1
run xor in1=+0.55 in2=-0.20 out=1
run xor in1=+1.16 in2=+0.79 out=0
run xor in1=-0.02 in2=-1.00 out=0
run xor in1=-1.32 in2=+1.83 out=1
run xor in1=+0.01 in2=+0.05 out=1
run xor in1=-0.25 in2=+0.97 out=1
run xor in1=-0.96 in2=+0.69 out=1
run xor in1=-1.18 in2=-2.04 out=0
run xor in1=+1.50 in2=+0.07 out=0
run xor in1=-0.53 in2=-0.79 out=0
run xor in1=-0.55 in2=-1.20 out=0
run xor in1=-1.04 in2=-0.19 out=0
run xor in1=-1.40 in2=+0.56 out=1
run xor in1=+0.05 in2=-0.83 out=1
run xor in1=+0.33 in2=-0.22 out=1
F1-score python=0.967741935483871 c=0.967741935483871
Total running time of the script: (0 minutes 0.537 seconds)