Note
Go to the end to download the full example code
Regression models
Simple demonstration of the different implemented regression models in emlearn
import os.path
import emlearn
import numpy
import pandas
import matplotlib.pyplot as plt
import seaborn
try:
# When executed as regular .py script
here = os.path.dirname(__file__)
except NameError:
# When executed as Jupyter notebook / Sphinx Gallery
here = os.getcwd()
Create dataset
Using a simple multi-class dataset included with scikit-learn
def load_dataset():
from sklearn import datasets
data = datasets.load_diabetes(as_frame=True)
df = data.data.copy()
df.columns = data.feature_names
df['target'] = data.target
return df
dataset = load_dataset()
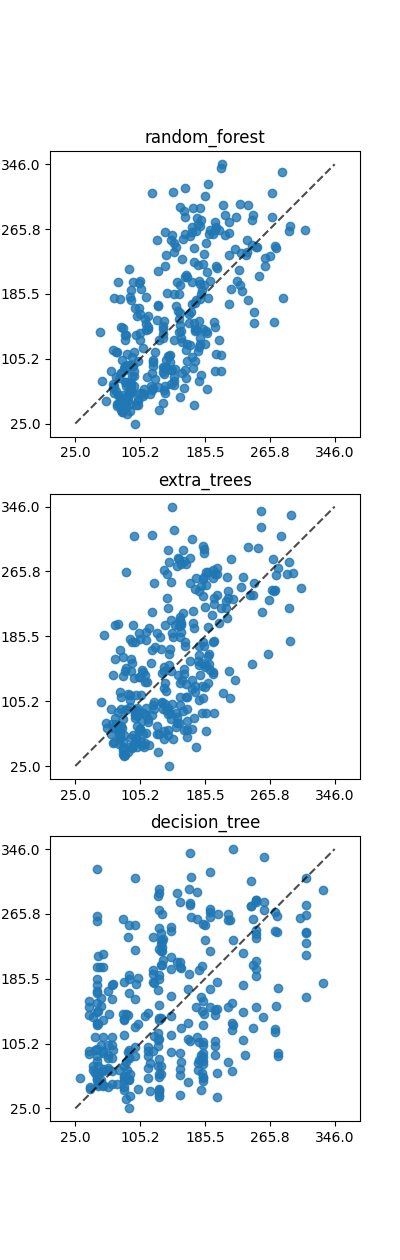
def plot_results(ax, model, X, y):
from sklearn.metrics import PredictionErrorDisplay
disp = PredictionErrorDisplay.from_estimator(
model, X, y,
ax=ax,
kind="actual_vs_predicted",
)
Train, convert and run model
Using the standard scikit-learn process, and then using emlearn to convert the model to C
def build_run_regressor(model, name, ax=None, feature_columns=None, color=None):
from sklearn.model_selection import train_test_split
target_column = 'target'
if feature_columns is None:
feature_columns = list(set(dataset.columns) - set([target_column]))
# Train model
test, train = train_test_split(dataset, test_size=0.3, random_state=3)
model.fit(train[feature_columns], train[target_column])
out_dir = os.path.join(here, 'regression')
if not os.path.exists(out_dir):
os.makedirs(out_dir)
model_filename = os.path.join(out_dir, f'{name}_model.h')
cmodel = emlearn.convert(model, method='loadable')
code = cmodel.save(file=model_filename, name='model')
test_pred = cmodel.predict(test[feature_columns])
# Generate a test dataet
test_data = numpy.array(test[feature_columns]).flatten()
test_res = numpy.array(test_pred).flatten()
# TODO: use tools to generate C file to compare classifier against expected results i C
if ax is not None:
plot_results(ax, model, X=test[feature_columns], y=test[target_column])
Run all models
Some of the supported modela and configurations
import sklearn.ensemble
import sklearn.tree
import sklearn.neural_network
import sklearn.naive_bayes
models = {
'random_forest': sklearn.ensemble.RandomForestRegressor(n_estimators=10, random_state=1),
'extra_trees': sklearn.ensemble.ExtraTreesRegressor(n_estimators=10, random_state=1),
'decision_tree': sklearn.tree.DecisionTreeRegressor(),
# TODO: support MultiLayerPerceptron
#'sklearn_mlp': sklearn.neural_network.MLPClassifier(hidden_layer_sizes=(10,10,), max_iter=30, random_state=1),
# TODO: support the various LinearRegression alternatives
}
# Based the below feature analysis - s5, bmi, bp are the most useful features for diabetes dataset
# https://scikit-learn.org/stable/auto_examples/ensemble/plot_gradient_boosting_regression.html#gsphx-glr-auto-examples-ensemble-plot-gradient-boosting-regression-py
feature_columns = ['s5', 'bmi', 'bp']
fig, axs = plt.subplots(
#ncols=len(models),
nrows=len(models),
figsize=(4, 4.2*len(models)),
sharex=False, sharey=False,
)
for model_no, (name, cls) in enumerate(models.items()):
ax = axs[model_no]
build_run_regressor(cls, name, ax)
ax.set_title(name)
ax.set_xlabel('')
plt.show()
Total running time of the script: (0 minutes 2.273 seconds)